Mapping transmission pathways of respiratory viruses in Kenya (SPReD-Kenya)
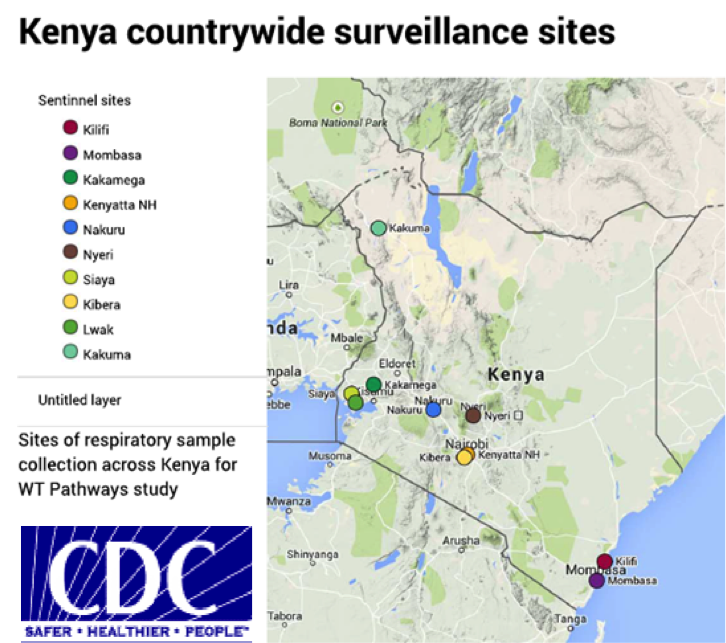
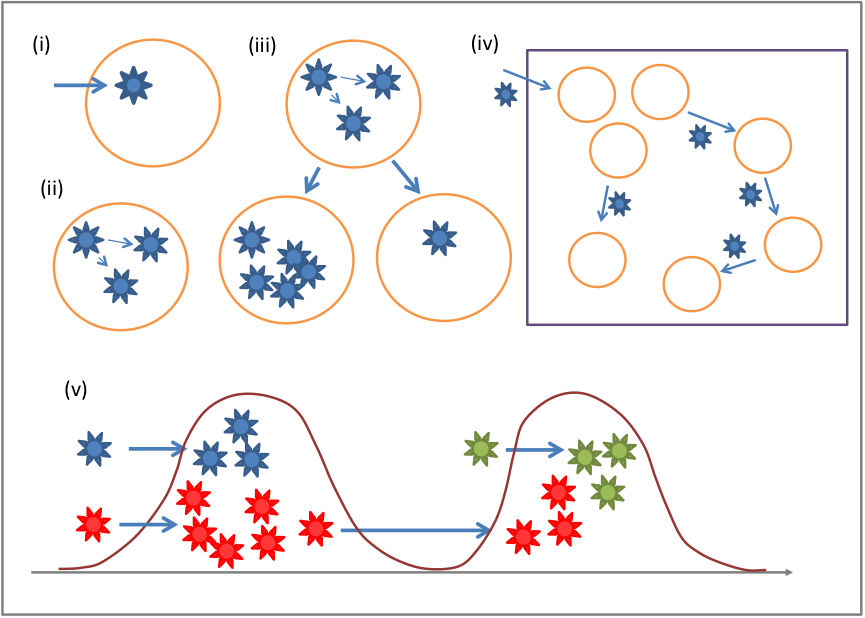
This is a collaborative project utilizing respiratory specimens and associated de-identified clinical data collected over a three-year period (2014-2016) from an existing influenza surveillance platform set up by KEMRI/CDC-Kenya. The aim is to define pathways of respiratory virus transmission, i.e. characteristic routes of virus introduction, spread, persistence and fade-out at different spatial and temporal scales (Figure 1), and to quantify transmission success (i.e. numbers of cases, extent of geographic spread, duration of persistence) and identify the factors that determine transmission potential. Approximately 7000 nasal specimens collected each year from ten influenza sentinel surveillance sites across Kenya from patients of various ages with severe acute respiratory illness (SARI) or influenza-like illness (ILI) will be analysed. Following initial rt-PCR screening, specimens positive for influenza viruses, RSV, human coronaviruses, rhinoviruses and other respiratory pathogens, will undergo whole genome deep sequencing. Resultant data will be used in both extensive phylogenetic and phylogeographic analyses and integrated mathematical models. The spatial-temporal case data combined with genetic finger-printing will enable us to understand how viruses spread within Kenya, and will ultimately inform on the most potent intervention modalities and locales. Besides its potential in informing policy on intervention strategies for respiratory viruses, the study will also improve on the current understanding on the characteristics of viral genetic diversity for the target pathogens in the Kenya setting.
Study PI: Patrick Munywoki (BSc, MSc,PhD)
Collaborators: Sandra dos Santos Chavez, Clayton Onyango, Jennifer Verani